These are the golden days of the Sequencing Era. The genome sequencing projects are so successfully transforming biomedical research that even former critics are now heralding the dawn of the Post-Sequencing Era. Plans are being made for high-throughput functional assays of all human genes, determination of all protein folds, and so on. However, sequencing isn't dead yet. Current investments in people and machines are such that sequencing will continue for some time at the world-wide rate of roughly one vertebrate genome per year. Studies like that of Gottgens et al. are desperately needed to bolster the meager factual basis for deciding which species to sequence next.
The advantages of the mouse as an experimental model and the deep understanding of mouse biology were so compelling that no other animal was given serious consideration as the second vertebrate to be sequenced. But which vertebrate will be the third? Among the candidates are puffer fish, chicken and rat, A species' utility for deciphering the human sequence may well be a pivotal consideration. Currently, there is very, very little hard data for determining which species would be most informative in this regard.
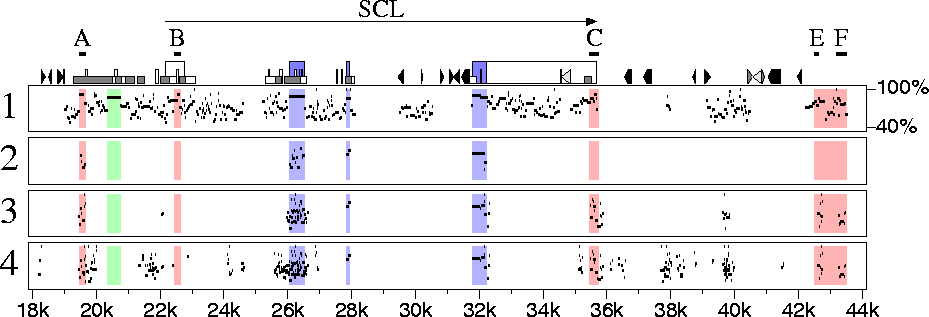
Gottgens et al. show that in one case, the SCL gene, a human-chicken sequence comparison adds much power to a human-mouse comparison for locating certain regulatory elements, but others are missed. Their analysis used three computer programs (dialign, dotter and BLAST) to compute alignments, and a fourth (the GCG package) to display some of the results. The results are robust, since an independent software system leads one immediately to similar conclusions (Figure 1). Regulatory elements labeled A, C, E and F stand out as conserved non-coding regions in the human-chicken comparison (panel 3), whereas element B is not identified as conserved at these program thresholds. Indeed, for SCL, the chicken sequence has advantages over the mouse sequence for comparison with human, since a large fraction of the matching sequences are implicated in function, either as protein coding regions (panel 2) or as potential regulatory elements (panel 3). In contrast, the human-mouse comparison (panel 1) shows many additional matches in sequences of undetermined function.

Figure 1. Percent identity plots of alignments between the human SCL gene and homologous regions from mouse and chicken. Features in the human sequence are shown along the top of box 1. Triangles indicate interspersed repeats, the shortest boxes are CpG islands and the tallest boxes are exons of SCL, with protein-coding regions colored blue. Red regions approximate those shown in Figure 2 of Gottgens et al. (Positions differ somewhat in the two figures, since here we use relative locations in the human sequence, while Gottgens et al. use positions along a particular human-mouse alignment.) Boxes labeled A, B, C, E and F correspond precisely to regions in Figure 3. (1) Alignments between the human and mouse sequences. (2) Alignments between the human and chicken sequences using the same thresholds as for panel 1. (3) Alignments between the human and chicken sequences using less stringent thresholds. (4) Alignments between the human and chicken sequences using the same thresholds as for panel 3, but not requiring that conserved regions be in the same order and orientations in the two species. The software used to prepare this figure can be run on user-supplied sequences on a server at the PipMaker website.
This gives us some hard data on utility of the chicken for explicating the human genome. However, these data come from only one gene, and our knowledge of its regulatory elements is incomplete. For example, does the region colored green in Figure 1 harbor a critical element? In the human-mouse comparison it stands out clearly as a 410-bp region with 87% nucleotide identity, but not a trace remains in the human-chicken comparison. Gottgens et al. earlier reported, that the region contains a DNaseI hypersensitive site and shows functional activity in transfection assays, which strongly suggests that it is functional in vivo. Thus, either the human-chicken comparison missed a region that is functional in mammals or the human-mouse comparison is leading us down a potentially expensive blind alley. I wonder which it is. Also, what is the situation with the other 80,000 or so human genes?
Assuredly, no one choice of the third species will solve all of our problems. While human-fish comparisons, may be good for revealing regulatory regions for Hox genes, even the chicken is at too great an evolutionary distance from humans for large-scale sequence comparisons to illuminate regulation of the beta-globin gene cluster, perhaps because the human and chicken clusters are not true orthologs. Mere phylogenetic distance is only one factor. For instance, consider the cardiac myosin heavy chain genes. The two genes, called alpha and beta, have different expression patterns in humans and mice. Mice express mainly the alpha form in their ventricles, whereas humans express almost exclusively the beta form. The difference depends on body size and, e.g., the pig more closely approximates the human expression pattern, perhaps making it better a candidate for identifying conserved regulatory signals in this region of the human genome. On the other hand, the alpha and beta genes are tandemly arrayed in a genomic region that is relatively similar between humans and mice, producing sequence matches in regions that are probably non-functional (as in panel 1 of Figure 1). Better differentiation between functional and non-functional regions might be obtained by using a species that is more distant from humans. Thus, optimal resolution of these genes' regulatory regions might be obtained by comparing the human sequence with sequence from a large, non-mammalian vertebrate, such as an ostrich, alligator or swordfish. This illustrates the points that (1) a variety of factors influence the utility of a particular species for comparative sequence analysis and (2) the best species varies with the gene.
The study by Gottgens et al. illuminates the choice of the third vertebrate. But time is running out. Within a very few years, decisions with huge fiscal ramifications may hinge on data of just this sort. We need a lot more of it to make an informed choice.